library(pdftools)
library(tidyverse)
library(ggthemes)
library(ggtext)
library(ggpubr)Another fun project – during my comprehensive exams I was given a large set of JCR papers to read and review. Since I had all these PDFs lying around, I had a good opportunity to learn more about automated data extraction from PDF documents. I decided to look at the distribution of P values in all 2019-2020 JCR papers.
Required Packages
Extract P Values
The code chunk below extracts the P values from papers in the following way:
- Get the names of all papers in a folder containing them
- Loop through each paper name and use the
pdftoolspackage to extract their raw text - Do some basic cleaning (e.g. getting rid of “\n”, which denote paragraph breaks)
- Extract all strings of text that are preceded by
"P >","P <","P =", and which end in a closed bracket")" - Output a data frame with 2 columns:
P valuesandPaper Name
files <- list.files("PDFs")
Results <- data.frame(matrix(ncol = 2, nrow = 0))
names(Results) <- c("P_Value", "Paper")
for (i in 1:length(files)) {
name <- files[i]
text <-
pdf_text(
here::here("blog", "jcr_pvals", "PDFs", name)
)
text <- gsub("\n", "", text)
text <- gsub(" ", "", text)
values <-
unlist(str_extract_all(text, 'p\\s?[=<>]\\s?\\.\\d{1,4}')) |>
as.data.frame() |>
mutate(Paper = name)
if (is_empty(values)) {
next
} else{
names(values) <- c("P_Value", "Paper")
Results <- rbind(values, Results)
}
}Cleaning
Now that I have every P value, I need to extract the actual number. This is moderately challenging - P values are often reported without the leading 0 (e.g. p = .07), and a p value that is reported as greater than 0.05 is different from one that is equal to or less than, and those differences need to be recorded somewhere for any future work I may do.
In summary, what the below code does is:
- Extract the “raw numeric value” from each reported p value string
- Replace the prior “p [<=>]” with a “0” instead
- Convert this column to numeric
Cleaned_Results <- Results |>
mutate(
Raw_Value = P_Value,
Raw_Value = gsub("p < ", "0", Raw_Value),
Raw_Value = gsub("p = ", "0", Raw_Value),
Raw_Value = gsub("p > ", "0", Raw_Value),
Raw_Value = gsub("p >", "0", Raw_Value),
Raw_Value = gsub("p =", "0", Raw_Value),
Raw_Value = gsub("p <", "0", Raw_Value),
Raw_Value = gsub("p< ", "0", Raw_Value),
Raw_Value = gsub("p= ", "0", Raw_Value),
Raw_Value = gsub("p> ", "0", Raw_Value),
Raw_Value = gsub("p<", "0", Raw_Value),
Raw_Value = gsub("p=", "0", Raw_Value),
Raw_Value = gsub("p>", "0", Raw_Value)
) |>
mutate(Operator = str_extract(P_Value, "[=<>]"))
Cleaned_Results$Raw_Value <- as.numeric(Cleaned_Results$Raw_Value)Plotting
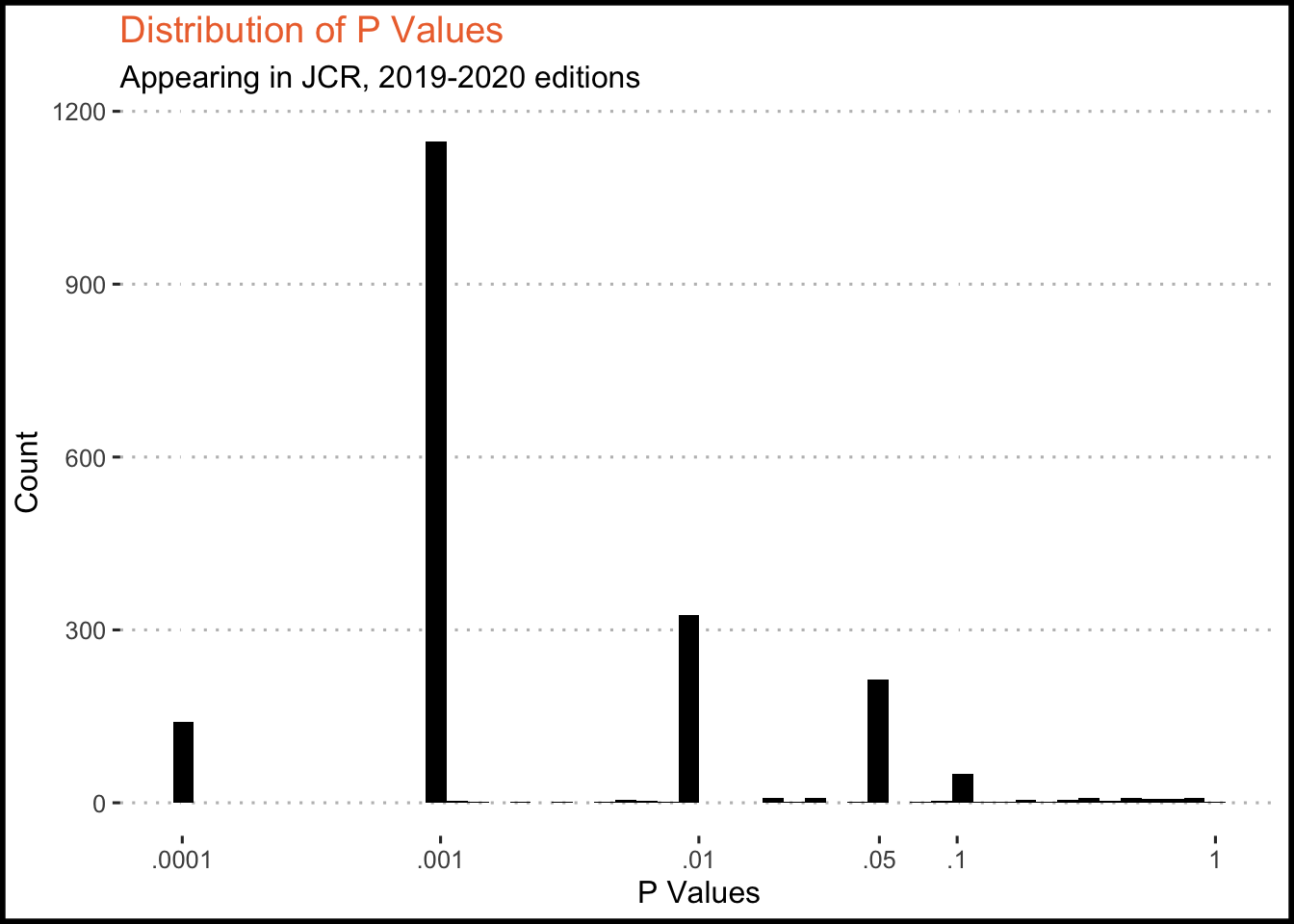
Finally, I wanted to plot the raw p values I’ve found. There’s little analytic code here - mostly just ggplot aesthetic wrangling. I used the {ggthemes} package for R to get the theme_fivethirtyeight function which gives me a lot of aesthetic power, for lack of a better term.
I’ve added X axis breaks at the standard p value thresholds - 0.1, 0.05, 0.01, 0.001. We should expect that there is significant clustering around these thresholds, as most researchers seem to report inequalities (p < x) rather than exact values.
Finally, p values aren’t exactly linear in their distribution, so everything is put on a log scale for easier interpretation.
ggplot(Cleaned_Results, aes(x = Raw_Value)) +
geom_histogram(bins = 50, fill = "black") +
scale_x_log10(
breaks = c(0.0001, 0.001, 0.01, 0.05, 0.1, 1),
labels = c(".0001", ".001", ".01", ".05", ".1", "1")
) +
theme_pubclean() +
theme(
axis.title.x = element_text(),
axis.title.y = element_text(),
plot.title = element_markdown(),
plot.background = element_rect(
colour = "black",
fill = NA,
size = 2
)
) +
labs(
title = "<span style = 'color: #ed713a;'>Distribution of P Values</span>",
subtitle = "Appearing in JCR, 2019-2020 editions",
x = "P Values",
y = "Count"
)